DF-Analyses of Heritability with Double-Entry
Twin Data: Asymptotic Standard Errors and
Efficient Estimation
Hans-Peter Kohler* Joseph Lee Rodgers†
July 17, 2001
|
Abstract
In Kohler and Rodgers (2000) we establish the asymptotic distribution of
DeFries–Fulker (1985) regression estimates for heritability and shared environmental
influences with double-entry twin data. A simple formula to estimate the covariance
matrix of the coefficients in DF-regressions is provided, and applications to simulated
data and Danish twin data show that this method can substantially increase the
statistical power of the analyses. We also provide an ‘efficient DF-analysis’ that yields
more precise estimates when additional covariates are included among the explanatory
variables. Below we provide programs to implement these methods in STATA and
other programs.
Keywords: DF-analysis, twin studies
Back to [ Hans-Peter Kohler] [ Data and Programs]
|
Contents
1 Introduction
DeFries and Fulker (1985) propose a method of estimating heritability (h2) and common
environmental influences (c2) with twin data by a simple linear regression of a twin’s trait on her
co-twin’s trait and the degree of genetic relatedness. The ambiguity in unselected
samples as to which twin’s trait should be used as the dependent, and which as the
independent variable, is frequently resolved by using double-entry (Haggard 1958).
Each twin pair is entered twice in the data, and each member of a twin pair provides
once the dependent and once the explanatory variable. While the consistency of the
regression estimates for heritability and environmental influences is not affected by
double-entry, the standard errors of the coefficients are obviously biased. It has been
common practice to adjust these standard errors for the correct degrees of freedom. In
large samples this adjustment is achieved by multiplying the standard errors obtained
from the double-entry regression with an adjustment factor 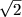 , or alternatively, by
multiplying the covariance matrix obtained in the initial regression by the factor 2.
However, this approach is problematic for at least two reasons. First, the DF-regression is
heteroscedastic and this potentially distorts the estimated covariance matrix of the parameters.
Second, the degrees-of-freedom adjustment achieved by multiplying covariance matrix
with a factor 2 is conservative. It assumes that double-entering a twin pair does not
increase the information—or loosely speaking, the overall degrees of freedom—in the
sample.
, or alternatively, by
multiplying the covariance matrix obtained in the initial regression by the factor 2.
However, this approach is problematic for at least two reasons. First, the DF-regression is
heteroscedastic and this potentially distorts the estimated covariance matrix of the parameters.
Second, the degrees-of-freedom adjustment achieved by multiplying covariance matrix
with a factor 2 is conservative. It assumes that double-entering a twin pair does not
increase the information—or loosely speaking, the overall degrees of freedom—in the
sample.
In order to overcome these limitations, we establish in Kohler and Rodgers (2000) the
asymptotically correct covariance matrix of the coefficients in DF-regressions with double-entry
data and we provide a simple estimator for this covariance matrix. The analysis is based
on the interpretation of DF-regressions as a generalized method of moment (GMM)
estimator. We also propose an ‘efficient DF-estimation’ that yields more accurate estimates
than DF-analysis when additional covariates are included among the right-hand-side
variables.
On this page we provide programs to implement the estimations proposed in Kohler and
Rodgers (2000). Primarily we will focus on STATA programs, but we will add alternative
programs as they become available (see Section 6). The programs provided below are subject to
our disclaimer.
2 Version and updates
This document is also available in pdf format. This html document and all other files
mentioned below are also available in a single zip archive ( dfregcv.zip).
- First version online: March 5, 2001
3 DF-Analysis: asymptotic standard errors and efficient estimation
The ‘augmented DF-analysis’ estimates the regression
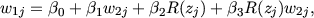 | (1) |
where wij is the trait value of twin i = 1, 2 in pair j, zj is the zygosity, and R(zj) is the degree
of genetic relatedness of the twin pair that depends on the zygosity zj. Unfortunately, when this
relation is estimated with double-entry data, the standard errors and variance-covariance matrix
provided by standard regression software are incorrect. Moreover, in Kohler and Rodgers
(2000) we also show that the commonly used degrees of freedom adjustment is not
satisfactory.
4 STATA programs for implementation
The estimation of the asymptotically correct standard errors provided in Result 1 of Kohler and
Rodgers (2000) can be implemented in STATA via three different pathways: (1) the robust
option of the regression command (regress); (2) the _robust command for computing
heteroscedasticity consistent variance-covariance estimators; and (3) the dfregcv program
described below.
The examples provided in this document replicate the analyses of body mass index in Section 5
of Kohler and Rodgers (2000).
4.1 Double entry twin data
The analyses are based on double-entry data. In the first entry of twin pair j in the data, the
trait of twin 1 is contained in a variable ‘traitvar1’ and that of twin 2 a variable ‘traitvar2’.
In the second entry of twin pair j in the data, traits are reversed. That is, in the second entry of
the twin pair the trait of twin 1 is contained in ‘traitvar2’ and that of twin 2 in
‘traitvar1’.
The data for the analyses of body mass index in Table 3 of Kohler and Rodgers (2000)
is an example of such double entry data. The variable names in this data set are:
. *! data description;
. describe twpair twentry birthy mono bmi* lbmi* ed_evsm* ed_amsm*;
twpair id of twin pair
twentry first or second entry of twpair
birthy Birth year of twin
mono dummy variable indicating MZ twin pairs
bmi1 body mass index, twin 1 or 2 (depending on twentry)
bmi2 body mass index, co-twin
lbmi1 log of BMI
lbmi2 log of BMI
lbmis1 log of BMI, age-pattern removed
lbmis2 log of BMI, age-pattern removed
ed_evsm within twin pair difference in ’ever smoked’
ed_amsm within twin pair difference in number of
cigarettes smoked
|
The first 10 rows of this data set look as follows. In these data, each twin pair occurs
twice and the assignment to ‘twin 1’ and ‘twin 2’ is reversed in the second entry.
. sort twpair twentry;
. list twpair twentry bmi* lbmis* in 1/10;
twpair twentry bmi1 bmi2 lbmis1 lbmis2
1. 116 1 26.85441 29.29688 .0665041 .153555
2. 116 2 29.29688 26.85441 .153555 .0665041
3. 149 1 21.61281 21.6041 -.1530994 -.1535023
4. 149 2 21.6041 21.61281 -.1535023 -.1530994
5. 151 1 22.34778 22.76944 -.108462 -.08977
6. 151 2 22.76944 22.34778 -.08977 -.108462
7. 157 1 23.8054 24.65483 -.0731012 -.0380408
8. 157 2 24.65483 23.8054 -.0380408 -.0731012
9. 162 1 18.02596 20.66116 -.3557666 -.219324
10. 162 2 20.66116 18.02596 -.219324 -.3557666
|
A straight forward possibility to implement the DF regression in Eq. (1) is via the regress
command as follows:
. *! implementation via standard OLS;
. *! a) generate degree of additive genetic relatedness;
. gen Radd = .5 + .5*(mono == 1);
. *! b) generate interaction between degree of genetic relatedness and;
. *! the co-twin’s trait value on the right-hand-side of the
> *! DF regression, where in our case this trait is contained in the
> *! variable ’lbmis2’;
. gen Rxtrait2 = Radd * lbmis2;
. *! c) perform DF regression;
. regress lbmis1 lbmis2 Radd Rxtrait2;
Source | SS df MS Number of obs = 1392
---------+------------------------------ F( 3, 1388) = 167.17
Model | 9.96705086 3 3.32235029 Prob > F = 0.0000
Residual | 27.585749 1388 .019874459 R-squared = 0.2654
---------+------------------------------ Adj R-squared = 0.2638
Total | 37.5527999 1391 .02699698 Root MSE = .14098
------------------------------------------------------------------------------
lbmis1 | Coef. Std. Err. t P>|t| [95% Conf. Interval]
---------+--------------------------------------------------------------------
lbmis2 | .0208506 .0697203 0.299 0.765 -.115918 .1576192
Radd | -.0003832 .0154114 -0.025 0.980 -.0306153 .029849
Rxtrait2 | .6664272 .0937798 7.106 0.000 .4824619 .8503926
_cons | .000303 .0114473 0.026 0.979 -.0221528 .0227589
------------------------------------------------------------------------------
|
While the coefficients in the above regression agree with those reported in Table 3 in Kohler and
Rodgers (2000), the standard errors reported by the STATA regression command are obviously
inappropriate because the data are double-entry.
In the next section we show, how the asymptotically correct standard errors can be estimated in
STATA (see Section 6 for implementations in programs other than STATA).
4.2 Estimation of standard errors
4.2.1 ‘robust’ option to regression command
The simplest possibility to correct the standard errors of the STATA regression command is to
use the robust cluster(twpair) option to the regress command. This estimation proceeds
almost identical to the regression in the previous section. The only modification is the option
robust cluster(twpair) that is added to the regress command:
. regress lbmis1 lbmis2 Radd Rxtrait2, robust cluster(twpair)
Regression with robust standard errors Number of obs = 1392
F( 3, 695) = 74.01
Prob > F = 0.0000
R-squared = 0.2654
Number of clusters (twpair) = 696 Root MSE = .14098
------------------------------------------------------------------------------
| Robust
lbmis1 | Coef. Std. Err. t P>|t| [95% Conf. Interval]
---------+--------------------------------------------------------------------
lbmis2 | .0208506 .1092156 0.191 0.849 -.1935814 .2352826
Radd | -.0003832 .0102665 -0.037 0.970 -.0205402 .0197738
Rxtrait2 | .6664272 .142898 4.664 0.000 .3858637 .9469908
_cons | .000303 .0090218 0.034 0.973 -.0174102 .0180163
------------------------------------------------------------------------------
|
The robust option specifies that robust standard errors (Newey and West 1987, White 1980)
instead of the usual OLS standard errors are calculated. The second part of the option,
cluster(twpair), additional specifies that the residuals of the regression are correlated within
twin pairs, and that only twin pairs—and not individuals—can be treated as independent
observations.
The above implementation, however, differs slightly from the standard errors specified in Result
1 of our paper. In particular, the above implementation via robust cluster(twpair) uses
n - k - 1, where n is the number of twin pairs and k is the number of right-and-side variables in
the regression, instead of n in the calculation of the standard errors according to
![a 1 ' -1 ' -1
^bDF - ® N (b,n-{E[xjxj]} L{E[xjxj]} ),](twdfeffprograms2x.gif) | (2) |
(see Result 1 in Kohler and Rodgers 2000). Asymptotically and in moderately large samples this
does not lead to noticeable differences, but in small samples it does make a difference. At the
moment, there does not seem to be a general rule as to whether n or n - k - 1 is more
appropriate in the calculation of standard errors in small samples (see also footnote 3 in our
paper).
4.2.2 ‘_robust’ command
In this subsection, we show how the default implementation of the robust cluster command in
STATA can be altered so that the standard errors are calculated exactly as given in our Result
1. This modification is based two steps: (a) a standard DF regression according to Eq. (1) that
yields correct coefficients but incorrect standard errors, and (b) a modification of the
variance-covariance matrix obtained in the initial regression with the _robust command. The
estimates are afterwards available for all STATA post-estimation commands such as test.
. quietly regress lbmis1 lbmis2 Radd Rxtrait2 if twpair != ., mse1;
* the ’if twpair != .’ avoids that twin pairs with a missing
* twin pair identifier (twpair) are included in the estimation;
. matrix D = e(V);
. mat b = e(b);
. predict double eps if twpair != ., residual;
. tab twentry if twpair != .;
* this tabulation gives the number of twin pairs, which
* equals 696 in our case;
first or |
second |
entry of |
twpair | Freq. Percent Cum.
------------+-----------------------------------
1 | 696 50.00 50.00
2 | 696 50.00 100.00
------------+-----------------------------------
Total | 1392 100.00
. estimates post b D, dof(696);
. _robust eps if twpair != ., minus(0) cluster(twpair);
. estimates display;
(standard errors adjusted for clustering on twpair)
------------------------------------------------------------------------------
| Robust
| Coef. Std. Err. t P>|t| [95% Conf. Interval]
---------+--------------------------------------------------------------------
lbmis2 | .0208506 .1090193 0.191 0.848 -.1931955 .2348968
Radd | -.0003832 .010248 -0.037 0.970 -.0205039 .0197375
Rxtrait2 | .6664272 .1426413 4.672 0.000 .3863685 .946486
_cons | .000303 .0090056 0.034 0.973 -.0173783 .0179844
------------------------------------------------------------------------------
|
The results obtained above agree exactly with the results reported in Table 3 of Kohler and
Rodgers (2000). Moreover, the differences between the above implementation and our earlier
estimation based on the robust cluster(twpair) option of the regress command
are very modest in our relatively large dataset and do not seem to be empirically
relevant.
4.2.3 The dfregcv command
The somewhat tedious estimation of the asymptotically correct standard errors in the previous
section can be avoided by using the dfregcv command. This is a user-written command,
and it allows the implementation of standard DF regressions and the efficient DF
regression with correct estimates of the standard errors. Moreover, this command
can also be used to combine DF regression with additional covariates that describe
non-shared environments (see Section 5.1 for a detailed description of the command
syntax).
The command syntax is simple. The first two variables after the dfregcv command give the
variables containing the trait values for twin 1 and 2 in the double-entry data. The subsequent
options define respectively the twin pair identifier (twpairv), the identifier for the first and
second entry of a twin pair (twentry), and the degree of genetic relatedness of DZ twins (rdz)
that equals .5 in additive genetic models, and .25 in dominance models. The final option
(dfmeth) specifies the method used for the estimation of the variance-covariance matrix and the
coefficients. In the example below, dfmeth(varest) specifies that the asymptotic
standard errors according to Result 1 in Kohler and Rodgers (2000) are calculated.
. dfregcv lbmis1 lbmis2, twpairv(twpair)
> twentry(twentry) mzdummy(mono)
> rdz(.5)
> dfmeth(varest);
**************************************************************
DOUBLE ENTRY DF ESTIMATION:
dependent variables: lbmis1 lbmis2
twin-pair identifier: twentry
MZ dummy: mono
genetic overlap DZ twins: .5
Method: varest
Covariates
Source | SS df MS # twin pairs = 696
---------+------------------------------ # twins (N) = 1392
Model | 9.96705086 4 2.49176271
Residual | 27.585749 1388 .019874459 R-squared = 0.2654
---------+------------------------------
Total | 37.5527999 1392 1
Asymptotically correct std. errors:
------------------------------------------------------------------------------
DFy | Coef. Std. Err. t P>|t| [95% Conf. Interval]
---------+--------------------------------------------------------------------
DFc2 | .0208506 .1090193 0.191 0.848 -.1930098 .234711
DFr | -.0003832 .010248 -0.037 0.970 -.0204865 .0197201
DFh2 | .6664272 .1426413 4.672 0.000 .3866115 .946243
DFcns | .000303 .0090056 0.034 0.973 -.017363 .0179691
------------------------------------------------------------------------------
|
The results of the command are displayed in a fashion that directly reveals the estimated h2 and
c2 of the DF analysis. The variables DFr and DFcns correspond to the degree of genetic
relatedness and the constant term in Eq. (1).
The results of this command exactly equals the results obtained by using the _robust command
in STATA in the previous section, and they agree exactly with the results in Table 3 of Kohler
and Rodgers (2000).
4.3 Combination with additional covariates
The dfregcv command is particularly convenient because it allows for an easy incorporation
of additional variables that describe differences in the non-shared environment of
twins.
In our data, we have used differences in smoking as important aspects in the non-shared
environment. These differences are described in the variables ed_evsm (difference in ever
smoked) and ed_evam (difference in the amount smoking).
The inclusion of these differences in non-shared environment via the dfregcv command simply
requires the addition of covars(ed_evsm ed_amsm) to the command syntax, where
ed_evsm and ed_amsm are variables describing differences in non-shared environments:
. *! inclusion of covariates;
. dfregcv lbmis1 lbmis2, twpairv(twpair)
> twentry(twentry) mzdummy(mono)
> rdz(.5)
> dfmeth(varest)
> covars(ed_evsm ed_amsm);
**************************************************************
DOUBLE ENTRY DF ESTIMATION:
dependent variables: lbmis1 lbmis2
twin-pair identifier: twentry
MZ dummy: mono
genetic overlap DZ twins: .5
Method: varest
Covariates
DFcov1 equals ed_evsm
DFcov2 equals ed_amsm
Source | SS df MS # twin pairs = 696
---------+------------------------------ # twins (N) = 1392
Model | 10.6750841 6 1.77918069
Residual | 26.8777157 1386 .019392291 R-squared = 0.2843
---------+------------------------------
Total | 37.5527999 1392 1
Asymptotically correct std. errors:
------------------------------------------------------------------------------
DFy | Coef. Std. Err. t P>|t| [95% Conf. Interval]
---------+--------------------------------------------------------------------
DFc2 | .0358214 .1084234 0.330 0.741 -.1768703 .2485132
DFr | -.0003725 .0099835 -0.037 0.970 -.0199569 .0192119
DFh2 | .6632649 .1380683 4.804 0.000 .3924195 .9341102
DFcov1 | -.0248875 .0097901 -2.542 0.011 -.0440925 -.0056824
DFcov2 | -.0018415 .0008847 -2.082 0.038 -.003577 -.0001061
DFcns | .0002954 .0088044 0.034 0.973 -.016976 .0175668
------------------------------------------------------------------------------
|
The header produced by the dfregcv command reports that the covariates have been named as
DFcov1 and DFcov2. The respective entries in the regression results reports the coefficients and
standard errors associated with these variables.
4.4 Efficient DF estimation
When additional covariates describing differences in non-shared environments are included in the
analysis, we have shown in Kohler and Rodgers (2000) that the above DF model is not efficient.
An improvement is feasible by using a GMM estimator that differentially weights the
observations contributed by MZ and DZ twins in the sample (see Section 3.2 in Kohler and
Rodgers 2000).
This efficient DF estimation is most easily implemented via the dfregcv command, and it only
requires the option dfmeth(gmm). This option specifies that the 2-step GMM estimator instead
of the standard DF regression is used in the analysis.
. *! efficient estimation;
. dfregcv lbmis1 lbmis2, twpairv(twpair)
> twentry(twentry) mzdummy(mono)
> rdz(.5)
> dfmeth(gmm)
> covars(ed_evsm ed_amsm);
**************************************************************
DOUBLE ENTRY DF ESTIMATION:
dependent variables: lbmis1 lbmis2
twin-pair identifier: twentry
MZ dummy: mono
genetic overlap DZ twins: .5
Method: gmm
Covariates
DFcov1 equals ed_evsm
DFcov2 equals ed_amsm
# twin pairs = 696
# twins (N) = 1392
Efficient GMM estimation:
------------------------------------------------------------------------------
DFy | Coef. Std. Err. z P>|z| [95% Conf. Interval]
---------+--------------------------------------------------------------------
DFc2 | .0159976 .1062916 0.151 0.880 -.1923302 .2243253
DFr | -.0007336 .0099766 -0.074 0.941 -.0202874 .0188202
DFh2 | .7192236 .1297151 5.545 0.000 .4649868 .9734605
DFcov1 | -.026052 .009765 -2.668 0.008 -.0451911 -.0069129
DFcov2 | -.0017482 .0007983 -2.190 0.029 -.0033129 -.0001836
DFcns | .000245 .0088043 0.028 0.978 -.017011 .0175011
------------------------------------------------------------------------------
|
5 Description of STATA programs
5.1 The dfregcv command
The dfregcv command is implemented via the dfregcv.ado file, and the corresponding help file
is dfregcv.hlp. These files need to be copied into the working directory (i.e., where the data file
is) or the personal directory for ado files. Moreover, the efficient DF analysis uses matginv for
the calculation of the pseudo-inverse, and the program matginv.ado needs to be added to this
directory (necessary only if these functions have not already been installed with STATA STB
dm49).
All files necessary for the implementation of dfregcv (including the help files and this document
in both html and pdf format) are included in dfregcv.zip.
Below is the help file (dfregcv.hlp) that describes the syntax of the dfregcv command in detail.
help for dfregcv
.-
DF Analyses with Double-Entry Twin Data: Asymptotic Standard
Errors and Efficient Estimation
-------------------------------------------------------------
dfregcv depvar1 depvar2 [if exp] [in range],
twpairv(varname) twentry(varname)
mzdummy(varname) rdz(#)
[dfmeth(string) covars(varlist) keepvar]
dfregcv shares the features of all estimation commands; see help
est (except, predict is not implemented).
Description
-----------
dfregcv performs DF analyses (DeFries and Fulker, 1985) with double
entry twin data, and (a) estimates the correct asymptotic standard
errors, (b) allows the combination of DF analyses with variables
describing observed differences in non-shared environments, and (c)
implements the efficient DF analysis via GMM estimation.
The data set needs to be in a "wide" form so that the information for
a twin pair is in one line of the dataset. Moreover, variables that
contain individual-specific information (e.g., education, weight,
etc.) need to be indicated with a suffix 1 or 2 for twin 1 and 2 in a
pair. Moreover, the data need to be double-entry, and each twin pair
thus needs to be included a second time with the twin assignment
reversed.
The variables depvar1 and depvar2 contain the phenotype of
interest.
See http://user.demogr.mpg.de/kohler for further examples.
Options for dfregcv
--------------------
twpairv(varname) specifies an unique identifier for each twin pair.
twentry(varname) specifies whether a specific line in a dataset is
the first or second entry of a twin pair (i.e., the variable
specified by twentry(varname) must either equal 1 or 2).
mzdummy(varname) specifies a dummy for MZ twins, i.e., it
specifies a variable that equals 1 for MZ and 0 for DZ twins.
rdz(#) specifies the genetic relatedness of DZ twins (which equals
.5 in standard models with additive genetic heritability).
dfmeth(string) specifies the method of estimation. varest
specifies that the asymptotically correct variance-covariance
matrix of DF-analysis is estimated (Result 1 in Kohler and
Rodgers, 2001). gmm specifies efficient DF estimation
(Result 2 in Kohler and Rodgers, 2001). times2 specifies a
degrees of freedom adjustment (i.e., multiplication of
standard errors with sqrt(2)). noadjust specifies that no
adjustment for double entry twin data is taken.
keepvar specifies that various variables generated by the program
dfregcv (all these variables are named DF*) are not deleted at
the end of the program. This is useful for parameter tests or
for further explorations of the data.
Examples
--------
* estimation of correct standard errors in DF analysis
.dfregcv lbmis1 lbmis2, twpairv(twpair)
twentry(twentry) mzdummy(mono)
rdz(.5)
dfmeth(varest)
* inclusion of additional variables describing differences
* in non-shared environment
.dfregcv lbmis1 lbmis2, twpairv(twpair)
twentry(twentry) mzdummy(mono)
rdz(.5)
dfmeth(varest)
covars(ed_evsm ed_amsm)
* efficient estimation of the above model
.dfregcv lbmis1 lbmis2, twpairv(twpair)
twentry(twentry) mzdummy(mono)
rdz(.5)
dfmeth(gmm)
covars(ed_evsm ed_amsm)
Note: in the above example, the variables lbmis1 and lbmis2 are
the logarithm of the body-mass-index for twin 1 and 2,
twentry is an identifier of twin pairs, twentry indicates the
first and second entry of a twin pair and ed_evsm and ed_amsm
describe differences in the smoking behavior between twins (see
http://user.demogr.mpg.de/kohler for further details).
|
6 Implementation with other programs
[yet to be written]
Back to [ Hans-Peter Kohler] [ Data and Programs]
References
DeFries, J. C. and D. W. Fulker (1985). Multiple regression analysis of twin data.
Behavior Genetics 15(5), 467–73.
Haggard, E. A. (1958). Intraclass Correlations and the Analysis of Variance. New York:
Dryden Press.
Kohler, H.-P. and J. L. Rodgers (2000). DF-analyses of heritability with double-entry
twin data: Asymptotic standard errors and efficient estimation. Behavior
Genetics 31(2), 179–191.
Newey, W. and K. West (1987). A simple positive semi-definite, heteroscadisticity and
autocorrelation consistent covariance matrix. Econometrica 55, 703–708.
White, H. (1980). A heteroscedasticity-consistent covariance matrix estimator and a
direct test for heteroscedasticity. Econometrica 48(4), 817–838.
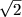 , or alternatively, by
multiplying the covariance matrix obtained in the initial regression by the factor 2.
However, this approach is problematic for at least two reasons. First, the DF-regression is
heteroscedastic and this potentially distorts the estimated covariance matrix of the parameters.
Second, the degrees-of-freedom adjustment achieved by multiplying covariance matrix
with a factor 2 is conservative. It assumes that double-entering a twin pair does not
increase the information—or loosely speaking, the overall degrees of freedom—in the
sample.
, or alternatively, by
multiplying the covariance matrix obtained in the initial regression by the factor 2.
However, this approach is problematic for at least two reasons. First, the DF-regression is
heteroscedastic and this potentially distorts the estimated covariance matrix of the parameters.
Second, the degrees-of-freedom adjustment achieved by multiplying covariance matrix
with a factor 2 is conservative. It assumes that double-entering a twin pair does not
increase the information—or loosely speaking, the overall degrees of freedom—in the
sample.
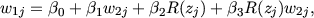
![a 1 ' -1 ' -1
^bDF - ® N (b,n-{E[xjxj]} L{E[xjxj]} ),](twdfeffprograms2x.gif)